Use Case: Linear Regression with Outlier Correction and Bootstrap
Andreas Brandmaier
2025-07-23
Source:vignettes/regression_example.Rmd
regression_example.RmdThis vignette demonstrates how to perform a robust linear regression analysis on the Boston housing data. We start by setting global chunk options and loading the required packages.
Load Data
We load the Boston housing data from the MASS package
and display the first rows to get an overview of the variables.
| crim | zn | indus | chas | nox | rm | age | dis | rad | tax | ptratio | black | lstat | medv |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.00632 | 18 | 2.31 | 0 | 0.538 | 6.575 | 65.2 | 4.0900 | 1 | 296 | 15.3 | 396.90 | 4.98 | 24.0 |
| 0.02731 | 0 | 7.07 | 0 | 0.469 | 6.421 | 78.9 | 4.9671 | 2 | 242 | 17.8 | 396.90 | 9.14 | 21.6 |
| 0.02729 | 0 | 7.07 | 0 | 0.469 | 7.185 | 61.1 | 4.9671 | 2 | 242 | 17.8 | 392.83 | 4.03 | 34.7 |
| 0.03237 | 0 | 2.18 | 0 | 0.458 | 6.998 | 45.8 | 6.0622 | 3 | 222 | 18.7 | 394.63 | 2.94 | 33.4 |
| 0.06905 | 0 | 2.18 | 0 | 0.458 | 7.147 | 54.2 | 6.0622 | 3 | 222 | 18.7 | 396.90 | 5.33 | 36.2 |
| 0.02985 | 0 | 2.18 | 0 | 0.458 | 6.430 | 58.7 | 6.0622 | 3 | 222 | 18.7 | 394.12 | 5.21 | 28.7 |
Code Chunk Reproduction Report
- ✅boston: REPRODUCTION SUCCESSFUL
Outlier Correction
Next, we remove observations with extremely high or low values of
medv using the IQR rule to reduce the influence of
outliers.
iqr_medv <- IQR(boston$medv)
q <- quantile(boston$medv, c(0.25, 0.75))
lower <- q[1] - 1.5 * iqr_medv
upper <- q[2] + 1.5 * iqr_medv
boston <- boston[boston$medv >= lower & boston$medv <= upper, ]Code Chunk Reproduction Report
✅iqr_medv: REPRODUCTION SUCCESSFUL
✅lower: REPRODUCTION SUCCESSFUL
✅q: REPRODUCTION SUCCESSFUL
✅upper: REPRODUCTION SUCCESSFUL
Linear Regression
We then fit a simple linear regression predicting median house value
from the percentage of lower status population (lstat).
model <- lm(medv ~ lstat, data = boston)
summary(model)
#>
#> Call:
#> lm(formula = medv ~ lstat, data = boston)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -12.6960 -2.6909 -0.7157 1.9776 14.6744
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) 30.25803 0.41087 73.64 <2e-16 ***
#> lstat -0.71853 0.02744 -26.19 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Residual standard error: 4.103 on 464 degrees of freedom
#> Multiple R-squared: 0.5964, Adjusted R-squared: 0.5956
#> F-statistic: 685.7 on 1 and 464 DF, p-value: < 2.2e-16Code Chunk Reproduction Report
- ✅model: REPRODUCTION SUCCESSFUL
Bootstrap Coefficients
To assess the variability of the coefficients we perform a simple bootstrap. Because analyses based on random numbers lead to different results across runs, we set a random seed to make this step reproducible. See Brandmaier et al. for a detailed discussion of reproducibility and random numbers (https://qcmb.psychopen.eu/index.php/qcmb/article/view/3763/3763.html).
set.seed(123)
boot_coefs <- replicate(1000, {
idx <- sample(nrow(boston), replace = TRUE)
coef(lm(medv ~ lstat, data = boston[idx, ]))
})
boot_means <- rowMeans(boot_coefs)
boot_means
#> (Intercept) lstat
#> 30.282183 -0.720951Code Chunk Reproduction Report
✅.Random.seed: REPRODUCTION SUCCESSFUL
✅boot_coefs: REPRODUCTION SUCCESSFUL
✅boot_means: REPRODUCTION SUCCESSFUL
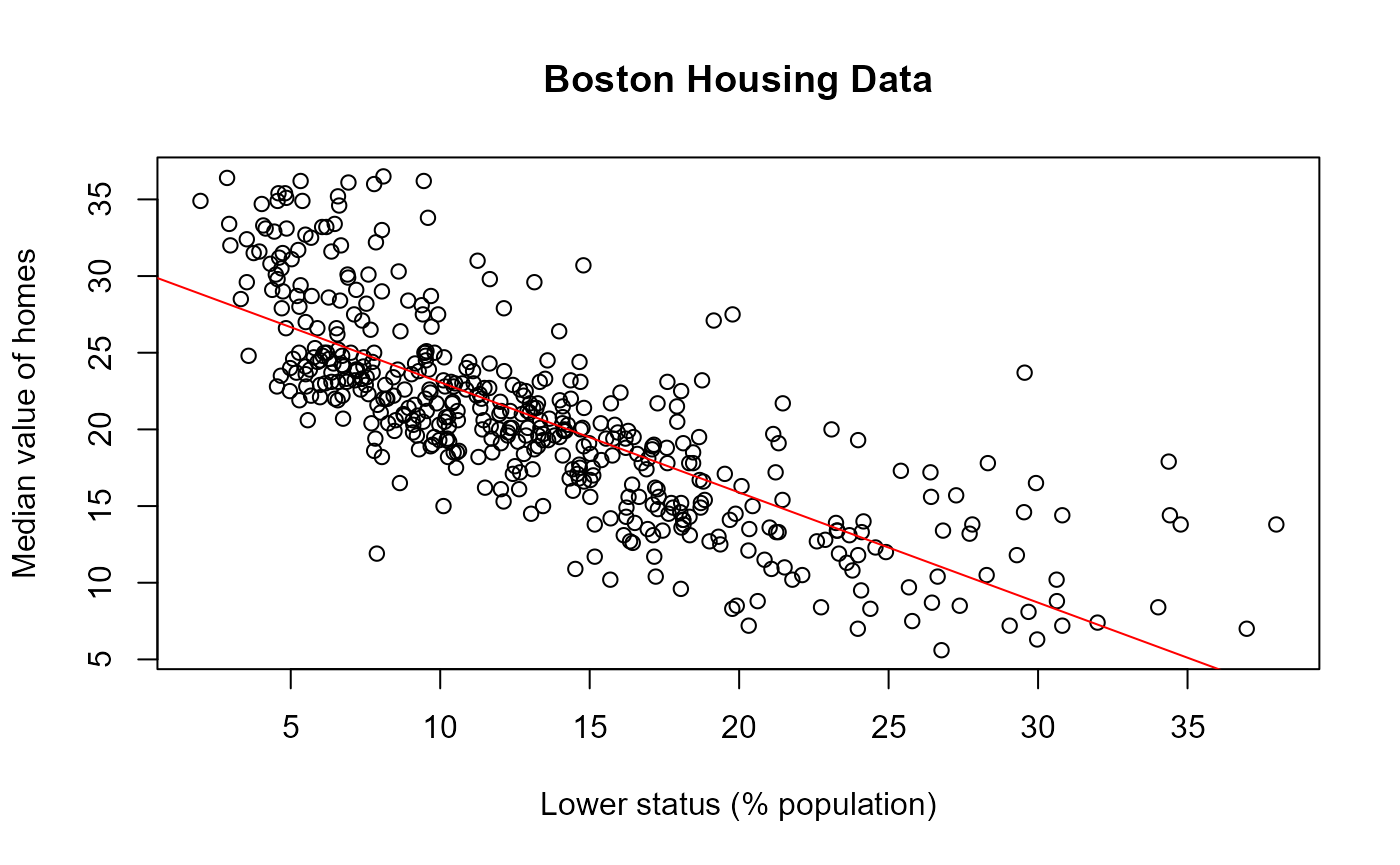
Plot
Finally, we visualise the relation between lstat and
medv together with the fitted regression line. Since plots
do return objects, they are not tested for reproducibility.
plot(boston$lstat, boston$medv,
xlab = "Lower status (% population)",
ylab = "Median value of homes",
main = "Boston Housing Data")
abline(model, col = "red")